ImageVU is a medical imaging repository that can be linked to clinical data through the Research Derivative or Synthetic Derivative.
ImageVU consists of magnetic resonance imaging (MRI) and computed tomography (CT), along with image metadata including study description, exam codes, and image acquisition date and time. Currently, ImageVU contains over 7.4 million MRI and CT image series, from 826,000 studies performed on 246,000 patients, dating back to 2007. Identified images are part of the Research Derivative (RD), which contains identified clinical data from across Vanderbilt. Images may also be de-identified and linked to the Synthetic Derivative (SD), which is a de-identified, HIPAA safe harbor clinical data repository.
Access
Images are made available by Integrated Data Access and Services Core (IDASC) as a billable service. Currently, users must go through IDASC in order to select and receive images. Metadata about the images is scheduled to be available in the RD Discover, SD Discover, and Record Counter self-service applications by the end of 2023. Investigators will then be able to use the RD or SD Discover self-service tool to output imaging accession numbers of interest or work with Integrated Data Access and Services Core (IDASC) to generate a list of accession numbers. Once a list of accession IDs has been obtained, IDASC’s ImageVU service can extract images and de-identify the DICOM files per project requirements as a billable service. To request an IDASC custom data pull or access to RD Discover or SD Discover, please fill out the common intake form, available here: https://redcap.vumc.org/surveys/?s=J8WNWKW7T4
Image De-identification
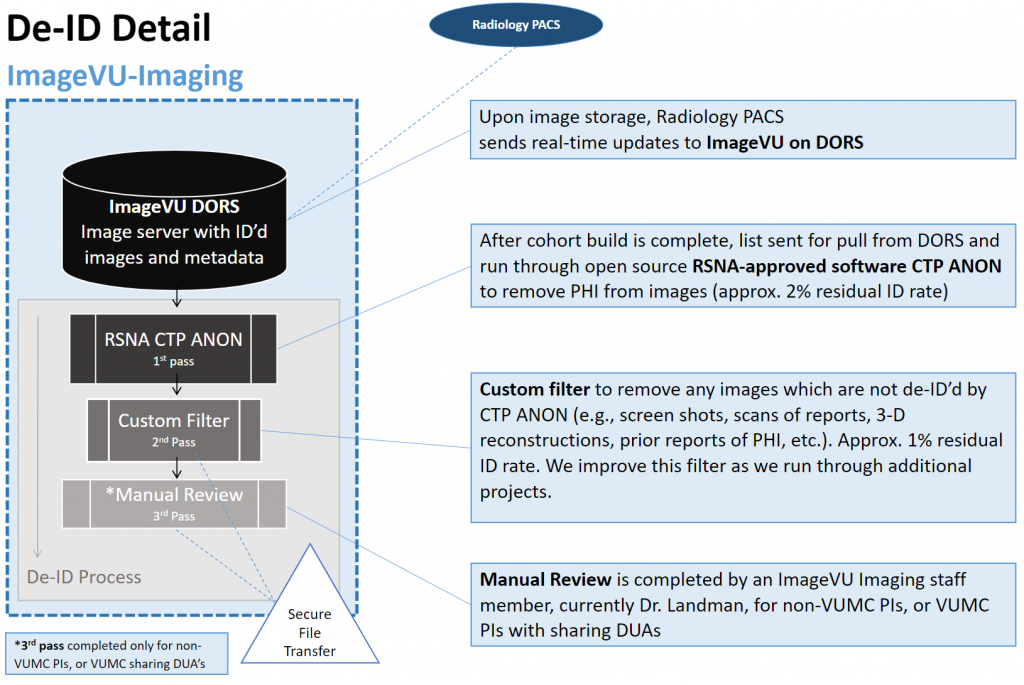
Identified images require an approved IRB study protocol, similar to any identified data. If de-identified images are requested, images will go through ImageVU’s two or three step de-identification process. ImageVU images can be de-identified on a per-project basis depending on the size of the project:
CTP ANON
Large projects (>1000 studies) are de-identified by first pushing them through a customized version of the RSNA’s CTP ANON software, then running them through custom Python scripts. Images that will be shared externally are then manually reviewed for PHI. Any residual PHI is noted and removed, and these findings are used to further customize and improve the Python scripts. The full three step process is required for any images that will be shared externally (outside of VUMC). The de-identification process consists of:
- Images are pushed through CTP ANON, an open source RSNA-approved software, to remove PHI from images. This step has a known residual PHI rate of about 2%.
- Custom filters are then applied to remove any images which are known to not be effectively de-identified by CTP ANON. This includes screenshots, scans of reports, 3-D reconstructions, and other known issues. This filter is continuously improved with each new project, and has a known residual PHI rate of <1%.
- Once images have run through the first two steps, each image undergoes a manual review and any images with residual PHI are removed from the study set.
Ambra
Small projects (<1000 studies) are well-suited for the Ambra platform, which de-identifies images using a combination of a metadata anonymization script and Tensor Flow AI scripts to redact pixelated PHI. Images are manually reviewed prior to being shared externally to VUMC, and patterns of residual PHI are reported back to the software manufacturer for quality improvement.
The Research Derivative (RD)
The Research Derivative is a database of clinical and related data derived from the Vanderbilt University Medical Center’s (VUMC) clinical systems and restructured for research. Data is repurposed from VUMC’s enterprise data warehouse, which includes data from StarPanel, VPIMS, and ORMIS (Operating Room Management Information System), EPIC, Medipac, and HEO among others. The medical record number and other person identifiers are preserved within the database. Data types include reimbursement codes, clinical notes and documentation, nursing records, medication data, laboratory data, encounter and visit data, among others. Output may include structured data points, such as ICD 9 or 10 codes and encounter dates, semi-structured data such as laboratory tests and results, or unstructured data such as physician progress reports. The database is maintained by the Office of Research Informatics under the direction of Paul Harris, PhD.
The Synthetic Derivative (SD)
Leveraging biomedical informatics expertise at Vanderbilt, a “mirror image” of the electronic health record (EHR), was created and termed the Synthetic Derivative (SD). The SD is an organized collection of de-identified information extracted from EHRs with content changed by deletion or permutation of all identifiers contained within each record. The SD contains ~3.5 million individual records with clinical information for the past 20 years available in a searchable form. HIPAA identifiers have been scrubbed with an error rate of <3%. New clinical data are added to the database as they are created. The database is maintained by the Office of Research Informatics under the direction of Robert Carroll, PhD.
Questions? Please contact us at: VICTRBigData@vumc.org