BioVU Description
BioVU
BioVU is Vanderbilt’s biorepository of de-identified samples extracted from discarded blood collected during routine clinical testing and linked to de-identified medical records in the Synthetic Derivative. The goal of BioVU is to provide a resource to Vanderbilt investigators for studies of genotype-phenotype associations.
Planning for BioVU began in mid-2004 and the first samples were collected in February 2007. Prior to collecting DNA samples, all aspects of the BioVU project were extensively tested. BioVU now accrues 500-1000 samples per week, totaling more than 300,000 DNA samples as of January 2023. Vanderbilt clinic patients may sign the BioVU Consent Form if they wish to donate their excess blood samples, or not sign the form if they do not wish to participate.
Collected Samples
Samples are scanned via a custom-developed sample acceptance program that includes automated exclusion based on specific criteria. Manual exclusions include poor quality of the blood sample, insufficient volume of blood and/or an unreadable label on the sample tube. Automated exclusions include opt-out, no signed form documenting notification of the program, duplicate samples not targeted for replenishment, and random exclusion. Once a sample passes the necessary criteria, it is accepted by the program. Acceptance of a sample triggers the encryption program to assign a unique research ID number to the sample. The unique research ID is generated by a Secure Hash Algorithm (SHA-512, National Security Administration). SHA-512 generated a unique 128 character (512 bit) code that serves as the unique research ID that links the samples to the de-identified clinical data and resulting genetic data. We have validated that the original medical record number (input) cannot be regenerated from the unique research ID (output).
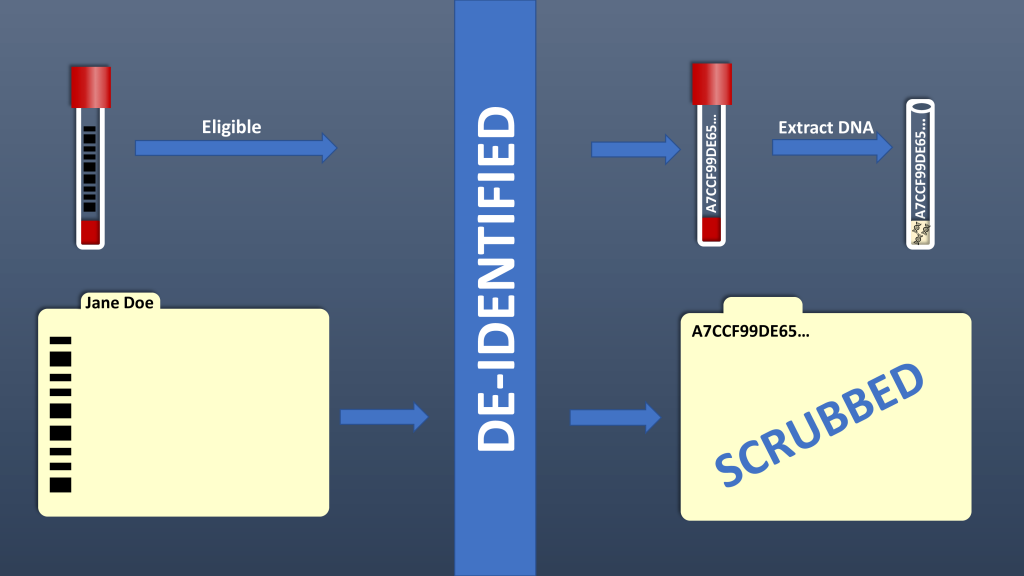
The SD
Leveraging biomedical informatics expertise at Vanderbilt, a “mirror image” of the electronic health record (EHR), was created and termed the Synthetic Derivative (SD). The SD is an organized collection of de-identified information extracted from EHRs with content changed by deletion or permutation of all identifiers contained within each record. The SD contains ~3.5 million individual records with clinical information for the past 20 years available in a searchable form. HIPAA identifiers have been scrubbed with an error rate of ~0.01%. New clinical data are added to the database as they are created. The records in the SD are labeled with the same 128-digit identifier as the sample to maintain the link between the clinical data and sample.
Usage
Samples may be requested after a detailed summary of the study is received, approved by the IRB and the Scientific Review Committee, and a user agreement is signed. A record counter tool, available to all Vanderbilt investigators and does not require IRB approval, can be used to estimate the number of cases and controls. Searches can be conducted using billing and procedure codes, free text searches in clinical notes (including histories, discharge summaries, laboratory reports, etc.), and laboratory results. The record counter searches can be filtered to estimate numbers of the cohort in the entire SD or only records associated with samples.
Initial examination of feasibility and utility of BioVU included assessment of sample handling algorithms and a “demonstration project”. The richness of phenotypic data was examined in a sample set of 26,724 records at the outset of the program. These records contained 6,816 unique ICD-9 codes and a total of 261,953 ICD-9 codes, with an average of 10 ICD-9 codes per patient. Natural language processing methods have been and continue to be developed to allow efficient generation of case and control cohorts.
BioVU served as a platform for Vanderbilt’s successful application to join NHGRI’s eMERGE (Electronic Medical Records and Genetics) Network. Vanderbilt is one of approximately 10 nodes in the network that link electronic medical records with genetic information to perform a series of studies examining the association between genetics and phenotypes extracted from the electronic medical record.
References:
1. Roden, D.M. et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clinical pharmacology and therapeutics 84, 362-9 (2008).
2. Pulley, J.M., Brace, M.M., Bernard, G.R. & Masys, D.R. Attitudes and perceptions of patients towards methods of establishing a DNA biobank. Cell and tissue banking 9, 55-65 (2008).
3. Pulley, J.M., Brace, M., Bernard, G.R. & Masys, D. Evaluation of the effectiveness of posters to provide information to patients about a DNA database and their opportunity to opt out. Cell and tissue banking 8, 233-41 (2007).
4. Pulley, J.M., Clayton, E., Bernard, G.R., Roden, D.M. & Masys, D.R. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci 3, 42-48 (2010).